Kiribati is a Pacific Island neighbour of Aotearoa New Zealand and has one of the highest rates of leprosy in the world. This stigmatised and neglected mycobacterial disease can cause major disability and is both treatable and preventable. With sea level rise threatening the existence of this low lying atoll, there is likely to be mass migration of i-Kiribati people to countries such as Aotearoa New Zealand in the future and thus a risk of reintroduction of this once-endemic disease.
Since 2018, the National Leprosy Programme in Kiribati has conducted screening and administration of annual single dose rifampicin chemoprophylaxis for two years for household contacts of newly diagnosed patients with leprosy. Despite this, the rate of leprosy remains extremely high. In 2021, the Ministry of Health and Medical Services in Kiribati (MH&MS) in collaboration with the University of Otago, the University of Sydney and the Pacific Leprosy Foundation (PLF), commenced a leprosy and tuberculosis mass screening and rifamycin-based chemoprophylaxis programme (PEARL + COMBINE) on the most populous island in Kiribati (population ~60,000). (1,2) Such largescale use of rifampicin has the potential to induce resistance in endemic Mycobacterium leprae strains which could have disastrous consequences for leprosy control.
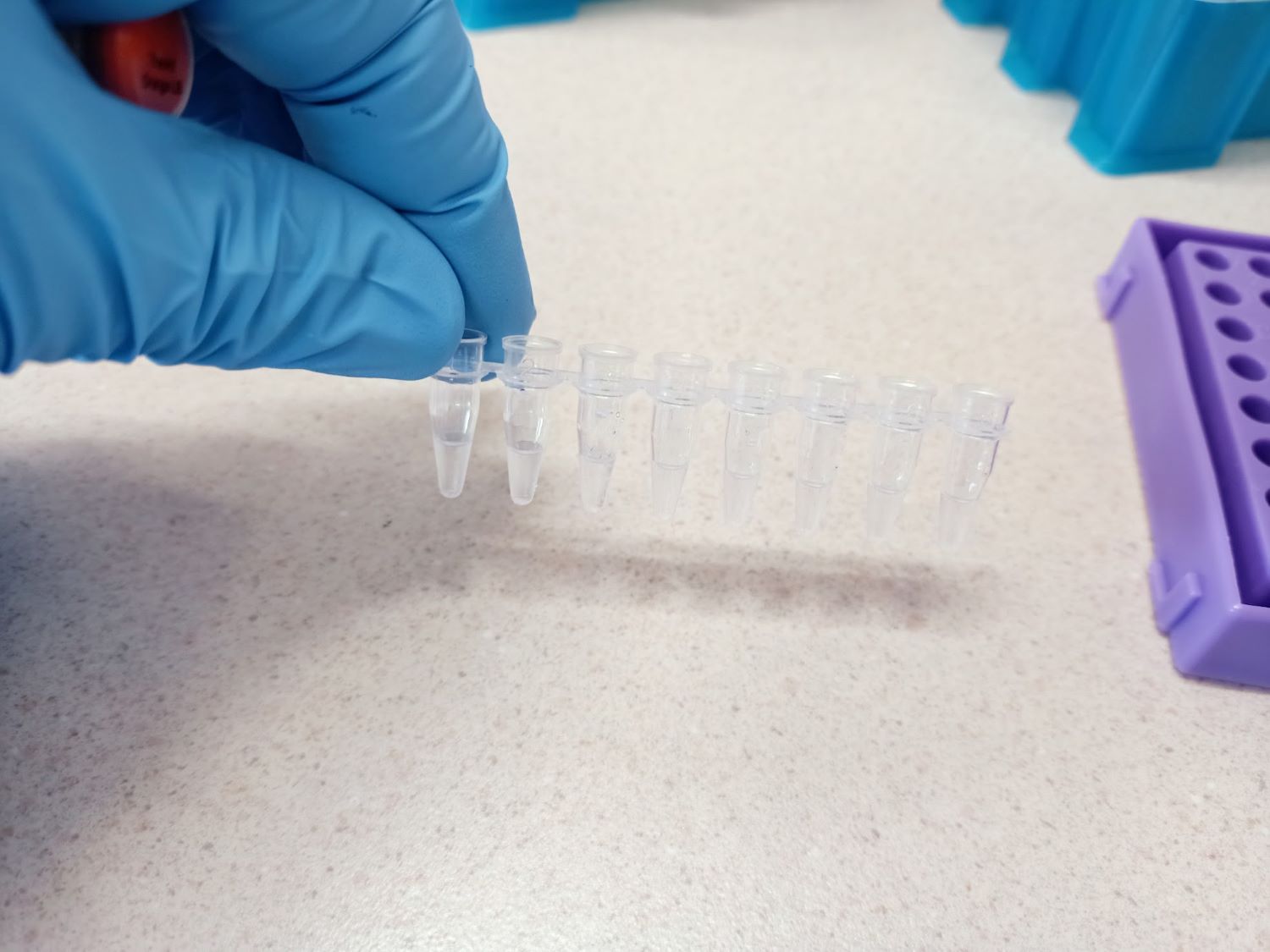
The researchers have previously set up a pilot M. leprae resistance monitoring programme for Kiribati isolates. This is technically complex as M. leprae cannot be cultured in vitro and testing needs to be done on M. lepraeDNA extracted from skin biopsies that have been transferred from Kiribati to Aotearoa New Zealand. Preliminary data, based on first generation methods, revealed baseline resistance rates to dapsone and rifampicin (both part of the standard leprosy treatment regimen) of 11% (9/83) and 0% respectively.
Currently, first-generation molecular testing relies on sequencing the rpoB, folP and gyrA genes extracted from skin biopsies of patients with newly diagnosed leprosy in Kiribati. Genoscreen France have developed a next generation deep sequencing technique (Deeplex Myc-Lep®) which targets drug resistance genes, SNPs and variable number of tandem repeats in M. leprae. This platform has been used in other leprosy endemic regions to survey drug resistance. Utilising multiplexed next generation deep sequencing, rather than current methods, should increase the sensitivity of drug-resistance testing, in particular due to additional coverage of parts of the genome recently identified as novel rifampicin resistance targets. The project involves conducting Deeplex sequencing on catalogued biopsy samples to enable comparison of the results of first-generation resistance testing. Researchers will also test prospectively acquired samples taken from patients with presumptive leprosy at the request of the MH&MS while the rifampicin prophylaxis project continues. This will provide valuable information on the impact of mass rifampicin prophylaxis on resistance patterns and will have the added benefit of helping to elucidate transmission pathways of resistant isolates.
A major concern for the leprosy treatment programme in Kiribati and all other endemic locations is treatment default (i.e. unplanned premature treatment cessation). Treatment default has major potential deleterious consequences including selection and transmission of drug resistant strains and inadequate treatment leading to development of irreversible deformity and disability. One of the major contributors to treatment default is severe adverse reactions to medication. There are two key factors that predispose patients to severe adverse reactions to dapsone, the prevalence of both of which is unknown in Kiribati.
The first of these is deficiency of glucose-6-phosphate dehydrogenase (G6PD - the most common enzymopathy globally) which predisposes to severe dapsone-induced haemolytic anaemia. The second is presence of the HLA-B*13:01 allele which predisposes to the potentially fatal dapsone hypersensitivity syndrome. This project will measure the prevalence of G6PD deficiency and the HLA-B*13:01 allele in the Kiribati population to help determine the utility of implementing pre-treatment screening of patients diagnosed with leprosy. Removal of dapsone from treatment regimens for patients who are positive for either condition should dramatically improve patient adherence and overall treatment effectiveness with consequent reduction in risk of resistance development.
The laboratory in Kiribati currently does not have capacity for G6PD testing, HLA-B13*01 allele detection or molecular detection of M. leprae or resistance-conferring mutations. A cornerstone of this work is to enhance laboratory capacity in Kiribati to enable in-country diagnostic testing to assist with clinical decision-making and potentially also local transmission hotspot investigation. The G6PD testing equipment used in the research (SD Biosensor®) lends itself well to bedside use in resource-poor settings and will facilitate ongoing use. Researchers also aim to begin preliminary development of a highly sensitive CRISPR molecular assay for M. leprae and drug resistance detection that would be suitable for use in Kiribati.